GWDataNoiseSource Tutorial¶
The GWDataNoiseSource is a data source element that generates realistic
gravitational wave detector noise with an appropriate power spectral density
(PSD). This tutorial will guide you through generating, analyzing, and
visualizing simulated gravitational wave strain data.
Overview¶
LIGO (Laser Interferometer Gravitational-Wave Observatory) and Virgo are gravitational wave detectors that measure tiny distortions in spacetime caused by passing gravitational waves. The background noise in these detectors has specific spectral characteristics.
The GWDataNoiseSource produces simulated strain data with noise characteristics inspired by modern GW detectors, useful for:
- Testing data analysis pipelines
- Developing and validating gravitational wave search algorithms
- Educational demonstrations
- Software validation
Step 1: Generate Simulated Strain Data¶
Let's create a script that generates simulated gravitational wave detector noise for multiple detectors:
#!/usr/bin/env python3
"""
Generate simulated LIGO/Virgo gravitational wave detector noise.
This script creates a pipeline with GWDataNoiseSource to generate strain
data for specified detectors and writes the output to text files.
"""
import os
import argparse
from sgn.apps import Pipeline
from sgnts.sinks import DumpSeriesSink
from sgnligo.sources.gwdata_noise_source import GWDataNoiseSource
def main():
# Parse command line arguments
parser = argparse.ArgumentParser(description="Generate gravitational wave detector noise")
parser.add_argument("--duration", type=float, default=50.0,
help="Duration in seconds (default: 50.0)")
parser.add_argument("--detectors", type=str, default="H1,L1",
help="Comma-separated list of detectors (default: 'H1,L1')")
parser.add_argument("--output-dir", type=str, default="./strain_output",
help="Directory to save output files (default: ./strain_output)")
parser.add_argument("--real-time", action="store_true",
help="Generate data in real time")
parser.add_argument("--verbose", action="store_true",
help="Print verbose output")
args = parser.parse_args()
# Create output directory
os.makedirs(args.output_dir, exist_ok=True)
# Parse detectors
detectors = [d.strip() for d in args.detectors.split(",")]
# Create channel dictionary
channel_dict = {detector: f"{detector}:FAKE-STRAIN" for detector in detectors}
# Set up pipeline
pipe = Pipeline()
# Create noise source
source = GWDataNoiseSource(
name="NoiseSource",
channel_dict=channel_dict,
duration=args.duration,
real_time=args.real_time,
verbose=args.verbose
)
# Create sinks for each channel
sinks = {}
for detector, channel_name in channel_dict.items():
output_file = os.path.join(args.output_dir, f"{detector}_strain.txt")
sinks[detector] = DumpSeriesSink(
name=f"Sink_{detector}",
sink_pad_names=[channel_name],
fname=output_file,
verbose=args.verbose
)
# Add source and sinks to pipeline
elements = [source] + list(sinks.values())
pipe.insert(*elements)
# Create link map
link_map = {}
for detector, channel_name in channel_dict.items():
link_map[f"Sink_{detector}:snk:{channel_name}"] = f"NoiseSource:src:{channel_name}"
# Add connections to pipeline
pipe.insert(link_map=link_map)
# Run the pipeline
print(f"Generating {args.duration} seconds of strain data for detectors: {detectors}")
pipe.run()
print("Pipeline completed successfully")
print(f"Output files written to {args.output_dir}")
if __name__ == "__main__":
main()
Save this script as generate_strain.py. You can run it with various options:
# Generate 50 seconds of data for LIGO Hanford (H1) and Livingston (L1)
python generate_strain.py
# Generate 30 seconds of data for all three detectors (H1, L1, V1)
python generate_strain.py --duration 30.0 --detectors H1,L1,V1
The script will create text files containing the simulated strain data, one file per detector.
Step 2: Analyze and Visualize the Data¶
Now let's create a script to analyze and visualize the simulated strain data:
#!/usr/bin/env python3
"""
Analyze and visualize simulated gravitational wave detector strain data.
This script reads strain data files, calculates spectral properties,
and creates visualizations including time series, PSDs, and spectrograms.
"""
import os
import argparse
import glob
import numpy as np
import matplotlib.pyplot as plt
from scipy import signal
def read_strain_file(filename):
"""Read strain data from a text file."""
# Load data, skipping any comment lines
data = np.loadtxt(filename, comments='#')
# Extract time and strain
times = data[:, 0]
strain = data[:, 1]
# Get detector from filename
detector = os.path.basename(filename).split("_")[0]
return times, strain, detector
def plot_strain_timeseries(times, strain, detector, output_dir):
"""Plot the strain time series data."""
# Make times relative to start
rel_times = times - times[0]
plt.figure(figsize=(12, 6))
# Plot time series
plt.plot(rel_times, strain, 'k-', linewidth=0.5)
plt.title(f"{detector} Strain Data Time Series")
plt.xlabel("Time (s)")
plt.ylabel("Strain")
plt.grid(True, alpha=0.3)
# Save figure
os.makedirs(output_dir, exist_ok=True)
plt.savefig(os.path.join(output_dir, f"{detector}_timeseries.png"), dpi=300)
plt.close()
def plot_strain_asd(strain, detector, output_dir, sample_rate=16384):
"""Calculate and plot the amplitude spectral density."""
# Calculate PSD using Welch's method
f, Pxx = signal.welch(strain, fs=sample_rate, nperseg=4096, noverlap=2048)
# Calculate ASD (amplitude spectral density)
asd = np.sqrt(Pxx)
plt.figure(figsize=(12, 6))
# Plot ASD
plt.loglog(f, asd)
plt.title(f"{detector} Amplitude Spectral Density")
plt.xlabel("Frequency (Hz)")
plt.ylabel("Strain/√Hz")
plt.xlim([10, sample_rate/2])
plt.grid(True, which="both", alpha=0.3)
# Save figure
os.makedirs(output_dir, exist_ok=True)
plt.savefig(os.path.join(output_dir, f"{detector}_asd.png"), dpi=300)
plt.close()
def plot_strain_spectrogram(strain, detector, output_dir, sample_rate=16384):
"""Create a spectrogram visualization."""
# Calculate spectrogram
f, t, Sxx = signal.spectrogram(strain, fs=sample_rate, nperseg=1024, noverlap=900)
plt.figure(figsize=(12, 6))
# Plot spectrogram
plt.pcolormesh(t, f, 10 * np.log10(Sxx), shading='gouraud')
plt.title(f"{detector} Strain Spectrogram")
plt.ylabel("Frequency (Hz)")
plt.xlabel("Time (s)")
plt.colorbar(label="PSD [dB/Hz]")
plt.yscale("log")
plt.ylim([20, sample_rate/2])
# Save figure
os.makedirs(output_dir, exist_ok=True)
plt.savefig(os.path.join(output_dir, f"{detector}_spectrogram.png"), dpi=300)
plt.close()
def create_detector_comparison(strain_files, output_dir, sample_rate=16384):
"""Create a comparison of ASDs from all detectors."""
plt.figure(figsize=(12, 8))
# Colors for each detector
colors = {"H1": "r", "L1": "b", "V1": "g"}
labels = {"H1": "LIGO Hanford", "L1": "LIGO Livingston", "V1": "Virgo"}
for file in strain_files:
times, strain, detector = read_strain_file(file)
# Calculate ASD
f, Pxx = signal.welch(strain, fs=sample_rate, nperseg=4096, noverlap=2048)
asd = np.sqrt(Pxx)
# Plot with detector-specific color and label
color = colors.get(detector, "k")
label = labels.get(detector, detector)
plt.loglog(f, asd, color=color, label=label, alpha=0.8)
plt.title("Detector Sensitivity Comparison")
plt.xlabel("Frequency (Hz)")
plt.ylabel("Strain/√Hz")
plt.xlim([10, sample_rate/2])
plt.grid(True, which="both", alpha=0.3)
plt.legend()
# Save figure
os.makedirs(output_dir, exist_ok=True)
plt.savefig(os.path.join(output_dir, "detector_comparison.png"), dpi=300)
plt.close()
def main():
parser = argparse.ArgumentParser(description="Analyze and plot strain data")
parser.add_argument("--input-dir", type=str, default="./strain_output",
help="Directory containing strain files (default: ./strain_output)")
parser.add_argument("--output-dir", type=str, default="./plots",
help="Directory to save plots (default: ./plots)")
args = parser.parse_args()
# Find all strain data files
strain_files = sorted(glob.glob(os.path.join(args.input_dir, "*_strain.txt")))
if not strain_files:
print(f"No strain data files found in {args.input_dir}")
return
print(f"Found {len(strain_files)} strain data files")
# Create output directory
os.makedirs(args.output_dir, exist_ok=True)
# Process each file
for file in strain_files:
print(f"Processing {file}")
times, strain, detector = read_strain_file(file)
# Create plots
plot_strain_timeseries(times, strain, detector, args.output_dir)
plot_strain_asd(strain, detector, args.output_dir)
plot_strain_spectrogram(strain, detector, args.output_dir)
# Create comparison plot
if len(strain_files) > 1:
create_detector_comparison(strain_files, args.output_dir)
print(f"Plots saved to {args.output_dir}")
if __name__ == "__main__":
main()
Save this script as analyze_strain.py. You can run it after generating strain data:
# Analyze and plot strain data
python analyze_strain.py --input-dir ./strain_output --output-dir ./plots
Example Output¶
Here's an example of what the generated strain plots should look like:
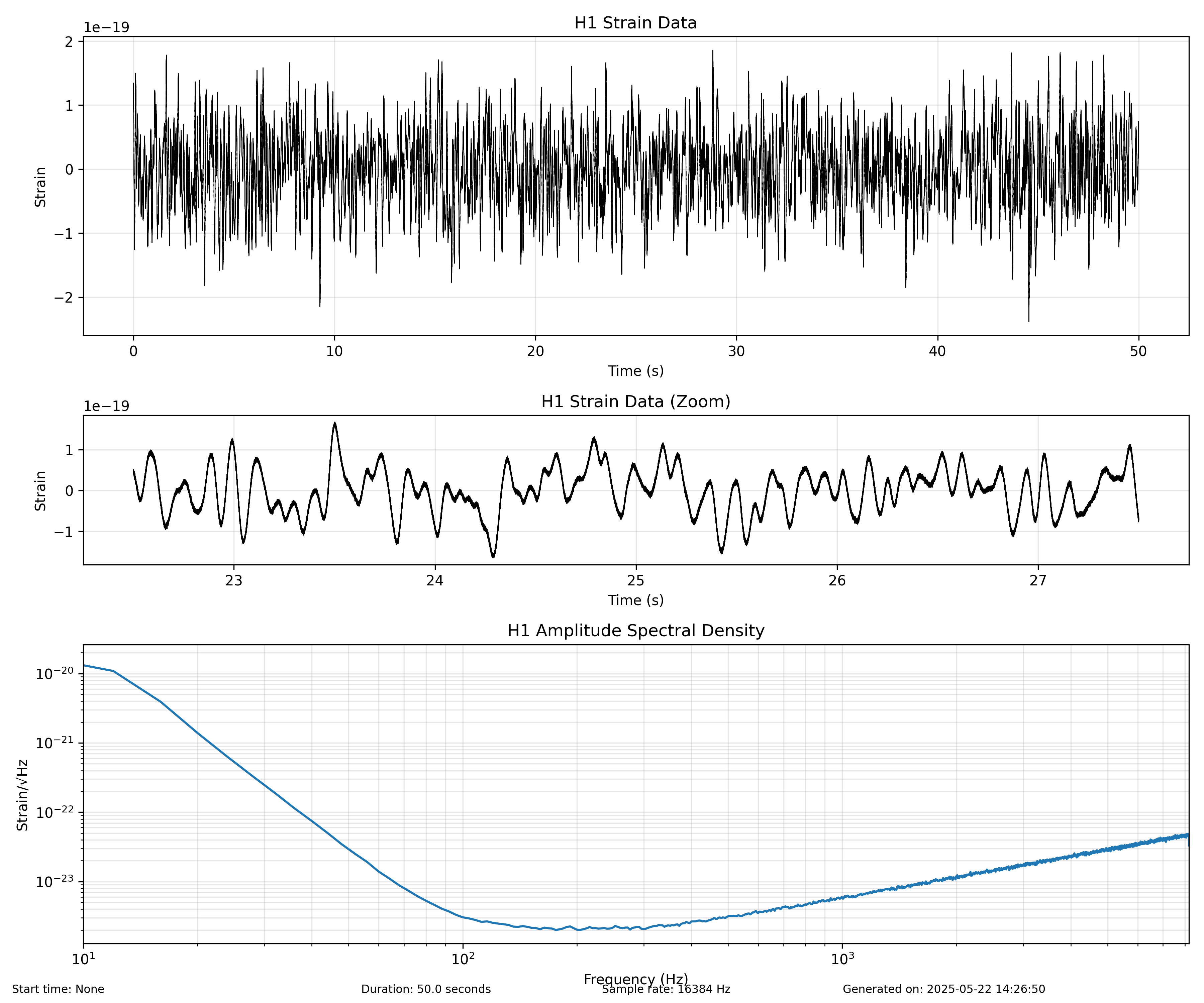
Figure: Example H1 strain data time series showing realistic gravitational wave detector noise with characteristic spectral properties.
GWDataNoiseSource Details¶
The GWDataNoiseSource has the following important parameters:
Key Parameters¶
name: Element name used in the pipelinechannel_dict: Dictionary mapping detector names to channel namest0: Start GPS time (defaults to current time if not specified)duration: Duration in secondsend: End GPS time (alternative to duration)real_time: If True, generates data in real timeverbose: If True, prints additional information
Supported Detectors¶
The source supports the following gravitational wave detectors:
H1: LIGO Hanford (Washington, USA)L1: LIGO Livingston (Louisiana, USA)V1: Virgo (Italy)
Each detector has a unique noise spectrum inspired by the sensitivity characteristics of the actual detectors.
Real-time Mode¶
For interactive applications, you can enable real-time mode:
# Example code:
source = GWDataNoiseSource(
name="NoiseSource",
channel_dict={"H1": "H1:FAKE-STRAIN"},
duration=60.0,
real_time=True, # Enable real-time generation
verbose=True
)
In real-time mode, the source will sleep between frames to maintain timing, simulating a live data feed.
Understanding the Output¶
The generated strain data has the following characteristics:
- Sample Rate: 16384 Hz (standard for LIGO/Virgo)
- Format: Two columns: GPS time and strain value
- Units: Strain is dimensionless (typically on the order of 10^-21 to 10^-22)
- Spectral Shape: Inspired by the sensitivity characteristics of each detector
The analysis scripts produce several visualizations:
- Time Series: Shows strain amplitude over time
- Amplitude Spectral Density (ASD): Shows the frequency-domain sensitivity
- Detector Comparison: Compares the ASD of multiple detectors
Conclusion¶
The GWDataNoiseSource provides a powerful way to generate realistic gravitational wave detector noise for testing and educational purposes. By using this source with the SGN pipeline framework, you can create sophisticated real-time or batch processing workflows for gravitational wave data analysis.
For more advanced applications, you can combine this noise source with other SGN elements to:
- Add simulated gravitational wave signals
- Apply whitening and filtering
- Perform matched filtering
- Develop and test detection algorithms